Environmental
Collaborating Scientist: Jayne Wu, Jiangang Chen, and Shigetoshi Eda
Project Abstract: Bisphenol A (BPA) is a high production volume chemical used in the production of polycarbonate plastics and epoxy resins for canned food. BPA enters surface water via effluent discharges during its manufacture, use and from waste landfill sites throughout United States. Fish among other aquatic species therefore have continuous, high exposure to BPA during their whole life cycle. BPA is an endocrine disruptor, the exposure to BPA in aquatic environment leads to broad negative ecological impacts including the altering of the development of male and female reproductive tracts in fish and effects are considered transgenerational. The field- measured BPA concentrations in water are critical for U.S. Environmental Protection Agency to develop water quality standards or environmental risk levels for the protection of human health. In this proposal, we will develop an aptamer-based, high sensitive and low cost microfluidic sensor for BPA detection. Compared to existing methods, our method will require no sophisticate instruments, low cost and suitable for real time onsite BPA surveillance to assist with the evaluation of ecological impact of BPA exposure.
Background and Significance: Recently, considerable interest has been focused on a wide variety of environmental endocrine disrupting substances (EDS). An EDS is a chemical with the potential to alter hormone action within the body. Several identified EDS are synthetic chemicals that have weak intrinsic hormonal or antihormonal activity, such compounds, when in the body, have the potential to interact with estrogen or androgen signaling mechanisms. This potential sex steroid bioactivity has drawn attention to the possible adverse effects of EDS on sex differentiation with specific emphasis on exposure during critical development. While research focuses and media reports are intensified with the impacts of EDS on human health, many EDSs such as DDT (dichlorodiphenyltrichloroethane) and PCBs (polychlorinated biphenyls) are banned or restricted primarily because they bioaccumulate and cause adverse effects on wildlife.
Bisphenol A (BPA) is a monomer widely used in a variety of consumer products, including number 7 plastics, food packaging, dental sealants, and thermal receipts. It was estimated that a total of 15 billion pounds was produced in BPA is a known endocrine disruptor with estrogenic, antiandrogen properties and can disrupt thyroid function. Studies have demonstrated that BPA can have significant biological effects at low, environmentally relevant doses. Because of the widespread use of BPA, the compound has been detected in aquatic environments, which are considered the ultimate reservoirs for environmental anthropogenic chemicals. In aquatic environment, fish among other aquatic species often has high exposure risk to BPA during entire life cycles including critical periods in development at the time that the pathological changes would likely be irreversible. BPA exposure disrupts gametogenesis in both male and female fish, alters development (neuronal, cardiac, germ cells, and sexual differentiation), changes sex ratios after embryonic exposure, induces mortality, and affects growth, time until first spawning, egg production, and fertilization success. In particular, there is concern that BPA enter in food chain and the consumption of fish could contribute to the total intake of BPA in human population.
To assess the ecological risk posed by chemicals in aquatic ecosystems, the field- measured BPA concentrations in water are critical for US Environmental Protection Agency to develop water quality standards or environmental risk levels for the protection of human health. Significant levels of BPA have been detected in river water, in river sediments in freshwater fish muscles. The majority of current BPA detection methods apply high performance liquid chromatography (HPLC) or enzyme-linked immunosorbent assay (ELISA), which requires the use of sophisticated and costly instruments by skilled personnel, and the operation is complex and time-consuming. In addition, the generation of antibody unavoidably requires to utilize animal host. Although good detection limit was
achieved by some less sophisticated methods, e.g. 1 pM by electrochemical aptasensor, they still require complicated experimental and data interpretation processes. Therefore, a simple and affordable method to monitor BPA level in environmental samples is highly desired.
Project Goals:
- Develop a high sensitivity, low cost aptamer based microfluidic sensor for BPA detection (sensor preparation and detection protocol).
- To optimize aptamer based capacitive microsensor for BPA detection in wastewater treatment plant effluent, and characterize the sensor performance including the limit of detection and specificity.
Collaborating Scientist: Kimberly Gwinn
Our discovery that colonization of the medicinal plant, echninacea, by mutualists enhances phytochemical content has significant implications for natural product industries and growers. Mycorrhizal plants produced up to 4.6-fold higher concentration of polyphenolics, and phytochemical content increased 30-fold. Medicinal alkylamides increased 1.7 fold in plants colonized with Beauveria bassiana, and up to 2.4-fold in plants colonized by both mycorrhizae and B. bassiana (Gualandi, M.S). Switchgrass extractives inhibited growth of foodborne and phytopathogenic bacteria both in vitro and in planta; this research resulted in a US patent (2016). Resin from Japanese umbrella pine, inhibited growth of some food spoilage and plant pathogenic bacteria, but stimulated growth of others (Yates, Ph.D.). The resin also inhibited growth of breast cancer cells. Chenopods (e.g., epazote, pitseed goosefoot, quinoa) produce a number of secondary metabolites that can be used as biopesticides. In tests with aqueous extracts of leaf material, antihelminthic activity, as measured by juvenile mortality and egg hatch, was correlated with saponin content except in epazote which produces a known antihelminthic. Catfish and corn stew cooked with pitseed goosefoot (one of the first domesticated crop species) had low populations of Bacillus cereus whereas high populations were found in stews cooked without the herb. This is the first evidence that the plant has antimicrobial activity and provides a plausible use for the crop (Goodman, Honors). High saponin cultivars of quinoa were less resistant to cucumber mosaic virus (CMV) and more resistant to nematodes. We have characterized a pathogen of hemp, Chaetomium globosum, and determined that plants with high concentrations of cannabinol (CBD) were more resistant to the pathogen than those with low CBD.
Fishes
Collaborating Scientists: Deb Miller and Mark Bevelhimer
Graduate Student: Ryan Saylor
Starting in 2018, I began working with my PI to secure separate funding to develop a realistic model fish with high bio-fidelity. I enjoy working with live fish but sought to create a biomimetic fish which may be as useful as a living organism. This project uses 3D printing technology, tissue simulating ballistic gelatin, and small sensors available at ORNL’s Manufacturing Demonstration Facility. I also used X-rays and CT scan data to quantify differences in skeletal architecture and other anatomorphic traits among riverine fishes. The latter was possible via collaboration with Radiologists and Veterinary Pathologists at the University of Tennessee’s Center for Veterinary Medicine. The model created (i.e., Gelfish) from this interdisciplinary collaboration will hopefully be used in place of live fish during trials associated with passage through hydropower turbines. We are still developing Gelfish and hope to include more sensors, higher sampling rates, and onboard data storage in future versions of this model.
 This image depicts our Gelfish model which is composed of ballistic gelatin designed to mimic the firmness of fish tissue. Gelfish also contains an embedded accelerometer to quantify the amount of force transferred to a fish’s body when impacted by hydropower turbine blades. This image represents the first generation of Gelfish technology. I am leading its continued development with help from experts at the Manufacturing Demonstration Facility at Oak Ridge National Laboratory.
This image depicts our Gelfish model which is composed of ballistic gelatin designed to mimic the firmness of fish tissue. Gelfish also contains an embedded accelerometer to quantify the amount of force transferred to a fish’s body when impacted by hydropower turbine blades. This image represents the first generation of Gelfish technology. I am leading its continued development with help from experts at the Manufacturing Demonstration Facility at Oak Ridge National Laboratory. The X-ray image above was taken by Dr. Silke Hecht and the Department of Radiology and Diagnostic Imaging at University of Tennessee Center for Veterinary Medicine. X-rays like this will allow me to quantify differences in vertebral morphometrics and assess how skeletal architecture may affect whole-body flexibility of riverine fishes struck by hydropower turbine blades. The top image is a white bass (related to striped bass) while the bottom is from a black redhorse (a species of sucker) – both are common to large rivers and reservoirs of eastern Tennessee.
The X-ray image above was taken by Dr. Silke Hecht and the Department of Radiology and Diagnostic Imaging at University of Tennessee Center for Veterinary Medicine. X-rays like this will allow me to quantify differences in vertebral morphometrics and assess how skeletal architecture may affect whole-body flexibility of riverine fishes struck by hydropower turbine blades. The top image is a white bass (related to striped bass) while the bottom is from a black redhorse (a species of sucker) – both are common to large rivers and reservoirs of eastern Tennessee.
Collaborating Scientists: Deb Miller, Mark Bevelhimer
Graduate Student: Ryan Saylor
A critical goal of DOE’s HydroPASSAGE project is to quantify the biological responses of fish to physical stressors like blade strike during passage through hydropower turbines. My specific role is to investigate how strikes from hydropower turbine blades affect rates of injury and mortality among riverine fishes. Controlled experiments are undertaken in the Aquatic Ecology Laboratory at ORNL in order to characterize biological responses in terms of mathematical models. More specifically, I generate dose-response relationships between blade strike velocity and mortality rates for a variety of fishes with different shapes and sizes. I also used novel multivariate statistical methods to analyze these data to better predict fish mortality. Finally, these data were used to parameterize software which is shared with turbine developers or dam owners/operators to better inform turbine design and increase passage survival of fishes.
Collaborating Scientist: Deb Miller
Graduate Student: Alison Cowie
Odontomas are benign tumors of dental tissue. In most animals, they are merely a nuisance, however in small fishes, such as the Ocellaris family, they can be deadly. About 50% of “fancy fin” Ocellaris develop odontomas, but the reasons are still unknown. Surgical removal is only possible with the larger bodied animals, so it is important to identify why they are forming. A possible genetic link has been proposed by researchers in Florida. It is the intent of this research to sequence the genetic code of both normal and fancy fin fishes, identify the breeding stock responsible, identify possible gene mutations, and provide histopathological evidence of gene expression.
 Ocellaris with odontoma.
Ocellaris with odontoma. Extracted odontoma from Ocellaris.
Extracted odontoma from Ocellaris.
Herpetology
Collaborating Scientists: Matt Gray, and Deb Miller
Batrachochytrium salamandrivorans (Bsal) is a fungal pathogen that likely originated in Asia, where local species harbor infections without signs of clinical disease. Bsal has recently been documented in Europe, where it is causing mass die-offs of some salamander species. It is hypothesized to have been introduced through unregulated international trade. To date, Bsal has not been detected in amphibian populations in North America. However, North America is a global hotspot for salamander biodiversity, accounting for 50 percent of species worldwide, so there is substantial concern about the possibility of Bsal’s introduction to the continent.
Collaborating Scientists: Matt Gray, Tim Sparer, and Rebecca Wilkes
Project Summary: Amphibian farming is a multi-billion dollar global industry that could be negatively impacted by highly virulent ranavirus strains. Moreover, international trade of amphibians infected with highly virulent strains could lead to spillover and mortality in wild populations, especially considering many captive facilities do not treat wastewater prior to discharge into the environment. The virulence trade-off hypothesis predicts that pathogen virulence will increase if transmission is guaranteed and there are abundant hosts. These conditions often exist in captive aquaculture facilities where fish and amphibian hosts are raised for human consumption or conservation. Several studies have shown that ranavirus isolates from captive facilities are more pathogenic than wild isolates; however, these studies compared one captive and one wild isolate (i.e., no replication). We tested for differences in virulence (as indexed by percent mortality and infection) among three ranavirus isolates from captive populations and three isolates from wild populations by performing controlled experimental challenges with tadpoles of two common amphibian host species (green frog, Lithobates clamitans; American bullfrog, L. catesbeianus). Infection and mortality were on average 30% greater for captive isolates compared to wild isolates for green frog tadpoles. Infection and mortality of bullfrog tadpoles were less than green frog tadpoles, and only one of the captive isolates was more pathogenic than the wild isolates. Our results provide preliminary evidence for evolution of increased virulence of ranaviruses in captive populations; however, the effects on host species are not uniform. Aquaculture facilities should consider routinely monitoring captive animals for ranavirus infection, and implementing strategies to thwart the evolution of pathogen virulence, such as reducing animal densities and quarantining or euthanizing infected individuals.
Impacts: The impacts for this study include:
- Co-housing amphibians at high densities in captivity can lead to increased virulence of ranavirus
- Virulent strains in captive amphibian populations can be translocated globally via international trade
- Ranaviruses that evolve in captivity are a threat to native e coastal species and thus may live in close proximity to feral cat colonies and sewage run-off
Results: This study documents for the first time that raising amphibians at high density in captivity can lead to the evolution of virulence in ranaviruses. These results have economic and conservation implications.
Collaborating Scientist: Deb Miller
Graduate Student: Ida Ayu Dian Kusuma Dewi
Climate change has been shown to impact many species worldwide. It causes significant problems such as altering marine nutrients and oxygen availability, raising sea levels, increasing ocean acidity, and increasing sea surface and beach temperature. Like many marine species, sea turtles are affected globally by climate change. Specifically, increasing beach temperatures are negatively affecting nesting populations. Hotter nest temperatures mean elevated incubation temperatures, and associated with these are sex ratios skewed towards feminization, reduced hatchling size, and lower hatchling success. In major nesting areas, elevated incubation temperatures are resulting in increased dead-in nest turtles. Along the Atlantic US, similar reduction in nest success occurs but without the precipitous declines seen in other populations. A recent study of the Atlantic leatherback turtle population along Florida USA (a less severely depleted population that serves as a model for the severely depleted populations elsewhere) suggests that increasing beach temperatures during incubation is negatively correlated to the intestinal length of the hatchlings. Shorter intestinal length is expected to reduce yolk absorption, and thus may potentially lower the survival rate of the hatchlings.
A major question generated from this study is whether this change equates to changes in the microanatomy, particularly the villi, which are necessary for nutrient absorption. Other questions include whether temperature also impacts hematopoietic tissue and lung development. These are fundamentally important to investigate as subsequent blood cell formation and oxygen perfusion and diving capabilities (of lungs) will determine survival of these active pelagic turtles. Among imperilled species, the leatherback is at risk of extinction, particularly in the Pacific, where poor nest success and illegal harvest of adults and eggs have led to declines in numbers. To date, very little is known regarding how climate change directly affects sea turtle nesting biology and its pre-hatchling development. This study will investigate the impacts of beach and incubation temperatures on hatchling GI structure, hematopoietic tissue formation, and lung development.
The goal of this novel research will be to provide the basis for assessing nest failures in order to prioritize nesting beach quality and identify mitigation measures as appropriate. Additionally, based on the data collected, this study will try to establish modelling of impact of temperature on Atlantic leatherback post hatchling growth and survival rate.
 A leatherback sea turtle hatchling found dead-in-nest.
A leatherback sea turtle hatchling found dead-in-nest. Early findings suggest that elevated temperature related to the shortened gastrointestinal tract of leatherback sea turtle hatchlings in Florida. Extending the study to microscopic measurement will provide us stronger basis to perform corrective reaction in improving nesting beach quality.
Early findings suggest that elevated temperature related to the shortened gastrointestinal tract of leatherback sea turtle hatchlings in Florida. Extending the study to microscopic measurement will provide us stronger basis to perform corrective reaction in improving nesting beach quality.
Collaborating Scientists: Rebecca Hardman, Deb Miller, Louise Rollins-Smith, Jhon Tailor Rengifo Mosquera, and Donald Walker
In collaboration with the Universidad Tecnológica del Chocó Diego Luis Córdoba, we have begun amphibian health surveys in towns surrounding the department capital of Quibdó. This is a pilot project to begin more intensive amphibian health research in a neglected region of Colombia, South America.
 Frog species collected for non-lethal sampling in the Colombian Chocó region.
Frog species collected for non-lethal sampling in the Colombian Chocó region. Frog species collected for non-lethal sampling in the Colombian Chocó region.
Frog species collected for non-lethal sampling in the Colombian Chocó region. Collaborators from the Universidad Tecnológica del Chocó Diego Luis Córdoba, performing herpetological surveys in disturbed forest.
Collaborators from the Universidad Tecnológica del Chocó Diego Luis Córdoba, performing herpetological surveys in disturbed forest. Collaborators from the Universidad Tecnológica del Chocó Diego Luis Córdoba, performing herpetological surveys in disturbed forest.
Collaborators from the Universidad Tecnológica del Chocó Diego Luis Córdoba, performing herpetological surveys in disturbed forest.
Collaborating Scientists: Rebecca Hardman, Deb Miller, Louise Rollins-Smith, Donald Walker, Matthew Niemiller, and TWRA
Under a collaborative state wildlife grant determined prevalence of amphibian pathogens and overall health of state threatened green salamander, Aneides aeneus. Performed molecular, microbiome, and histopathological analysis.
 TWRA biologists survey for Green Salamanders.
TWRA biologists survey for Green Salamanders. Adult green salamander with a unique pattern ID for recapture analysis.
Adult green salamander with a unique pattern ID for recapture analysis. Green salamanders in a sterile water rinse for skin peptide collection.
Green salamanders in a sterile water rinse for skin peptide collection.
Collaborating Scientists: Rebecca Hardman and Deb Miller
Evaluation of toe lesions and associated changes in skin peptides, microbiomes, and pathogen prevalence in wild populations. This is ongoing field and laboratory-based project.
 Previous UT vet student Kendall Oziminski and UT undergraduate Alexander Miel collect Hellbender skin secretions.
Previous UT vet student Kendall Oziminski and UT undergraduate Alexander Miel collect Hellbender skin secretions. Adult and juvenile hellbenders in sterile water bath.
Adult and juvenile hellbenders in sterile water bath. Dr. Hardman evaluates an adult hellbender.
Dr. Hardman evaluates an adult hellbender.
Collaborating Scientists: Rebecca Hardman, Deb Miller, and the Nashville Zoo at Grassmere
We are in our second phase of testing subcutaneous implants impregnated with terbinafine in juvenile hellbenders. This is a collaboration with the Nashville Zoo at Grassmere to prevent mortality due to chytridiomycosis in captive-raised juvenile hellbenders when released into the wild in 2021.
 Sherri Doro Reinsch, herpetologist at the Nashville Zoo at Grassmere and Dr. Hardman collecting blood from a juvenile hellbender. This collaborative study with the zoo looked at efficacy of subcutaneous implants as an efficient means for terbinafine administration without the need for daily handling.
Sherri Doro Reinsch, herpetologist at the Nashville Zoo at Grassmere and Dr. Hardman collecting blood from a juvenile hellbender. This collaborative study with the zoo looked at efficacy of subcutaneous implants as an efficient means for terbinafine administration without the need for daily handling.
Collaborating Scientists: Deb Miller and Rebecca Hardman
Continued investigation of several cases and mortality events of wild amphibian species throughout the state of Tennessee and abroad. We are using molecular and histopathological analyses to discover infectious agents that may be threatening wild populations.
Collaborating Scientists: Rebecca Hardman, William Sutton, and Donald Walker
Collaborating with state agencies, Tennessee State University, and Middle Tennessee University to perform histopathological confirmation of snake fungal disease due to Ophidiomyces ophiodiicola and provide veterinary expertise in coelomic implantation of transmitters in the state endangered pygmy rattlesnake (Sistrurus miliarius).
 Eastern Kingsnake with lesions consistent with snake fungal disease (SFD).
Eastern Kingsnake with lesions consistent with snake fungal disease (SFD). Pygmy Rattlesnake getting a drink of water upon recovery of transmitter implantation surgery.
Pygmy Rattlesnake getting a drink of water upon recovery of transmitter implantation surgery. TSU researcher Dr. Bill Sutton, getting ready to track one of the study pygmy rattlesnakes.
TSU researcher Dr. Bill Sutton, getting ready to track one of the study pygmy rattlesnakes.
Collaborating Scientists: Rebecca Hardman, William Sutton, and Michael Freake
Performing surgical implantation of radio transmitters in hellbenders for a collaborative eastern hellbender (Cryptobranchus alleganiensis alleganiensis) translocation project lead by collaborators from Tennessee State University and Lee University. We continue to oversee any subsequent field collection and laboratory processing of health samples of tracked individuals.
 Dr. Rebecca Hardman (UTK) surgically implanting a transmitter in an individual for post-translocation monitoring.
Dr. Rebecca Hardman (UTK) surgically implanting a transmitter in an individual for post-translocation monitoring. Project leaders Dr. Bill Sutton (Tennessee State University [TSU]) and Dr. Michael Freake (Lee University) with TSU graduate student Brad Nissen about to release an adult hellbender to its new stream.
Project leaders Dr. Bill Sutton (Tennessee State University [TSU]) and Dr. Michael Freake (Lee University) with TSU graduate student Brad Nissen about to release an adult hellbender to its new stream.
Collaborating Scientists: Matt Gray, Robert J. Ossiboff, Kurt Ash, and Debra Miller
Graduate Students: Anastasia Towe, Davis Carter, Markese Bohanon, and Brittany Bajo
Batrachochytrium salamandrivorans is an emerging fungus that is causing salamander declines in Europe. We evaluated whether an invasive frog species (Cuban tree frog, Osteopilus septentrionalis) that is found in international trade could be an asymptomatic carrier when exposed to zoospore doses known to infect salamanders. We discovered that Cuban tree frogs could be infected with Batrachochytrium salamandrivorans, and surprisingly that chytridiomycosis developed in animals at the two highest doses. To fulfill Koch’s postulates, we isolated Batrachochytrium salamandrivorans from infected frogs, exposed eastern newts (Notophthalmus viridescens) to the isolate, and verified infection and disease using histopathology. This experiment represents the first documentation of Batrachochytrium salamandrivorans chytridiomycosis in a frog species, and substantially expands the conservation threat and possible mobilization of this pathogen in trade.
 Cuban treefrogs showing representative lesions of Bsal chytridiomycosis, including erythema and hemorrhage on the feet and ventrum, excessive shedding on the feet, darkened pigmentation on the dorsum, and petechia.
Cuban treefrogs showing representative lesions of Bsal chytridiomycosis, including erythema and hemorrhage on the feet and ventrum, excessive shedding on the feet, darkened pigmentation on the dorsum, and petechia. Cuban treefrogs with Bsal lesions (color inversion).
Cuban treefrogs with Bsal lesions (color inversion).
Batrachochytrium dendrobatidis and B. salamandrivorans are important amphibian pathogens responsible for morbidity and mortality in free-ranging and captive frogs, salamanders, and caecilians. While B. dendrobatidis has a widespread global distribution, B. salamandrivorans has only been detected in amphibians in Asia and Europe. Although molecular detection methods for these fungi are well-characterized, differentiation of the morphologically similar organisms in the tissues of affected amphibians is incredibly difficult. Moreover, an accurate tool to identify and differentiate Batrachochytrium in affected amphibian tissues is essential for a specific diagnosis of the causative agent in chytridiomycosis cases. To address this need, an automated dual-plex chromogenic RNAScope® in situ hybridization (ISH) assay was developed and characterized for simultaneous detection and differentiation of B. dendrobatidis and B. salamandrivorans. The assay, utilizing double Z target probe pairs designed to hybridize to 28S rRNA sequences, was specific for the identification of both organisms in culture and in formalin-fixed paraffin-embedded amphibian tissues. The assay successfully identified organisms in tissue samples from five salamander and one frog species preserved in formalin for up to 364 days and was sensitive for the detection of Batrachochytrium in animals with qPCR loads as low as 1.1 × 102 zoospores/microliter. ISH staining of B. salamandrivorans also highlighted the infection of dermal cutaneous glands, a feature not observed in amphibian B. dendrobatidis cases and which may play an important role in B. salamandrivorans pathogenesis in salamanders. The developed ISH assay will benefit both amphibian chytridiomycosis surveillance projects and pathogenesis studies by providing a reliable tool for Batrachochytrium differentiation in tissues.
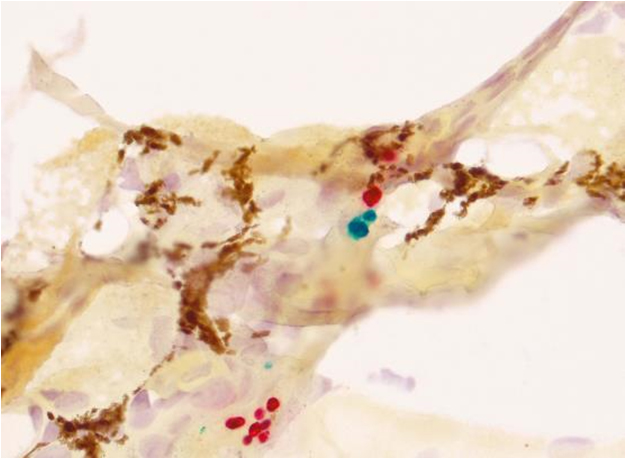 Histological samples from a Bd and Bsal co-infected newt, stained with ISH protocol resulting in Bd organisms in green and Bsal organisms in red.
Histological samples from a Bd and Bsal co-infected newt, stained with ISH protocol resulting in Bd organisms in green and Bsal organisms in red.
Batrachochytrium salamandrivorans (Bsal) is a recently discovered fungal pathogen that is causing salamander declines in Europe. Bsal has two life forms: a motile, flagellated zoospore and a non-motile, highly adhesive encysted spore, which differ in their environmental persistence. Our goal was to optimize flow cytometry to differentiate between viable and dead zoospores as well as encysted and flagellated spore types. Understanding the shedding rate of viable zoospores for each life form is fundamental to modeling Bsal epidemiology and predicting likelihood of emergence and geographic spread. We optimized flow cytometry for Bsal enumeration then compared the variability in counts between this technology and traditional hemocytometer counts. Hemocytometer counts were more variable than flow cytometry counts for researchers with limited to moderate hemocytometer experience, but not for experienced researchers. We optimized flow cytometry to differentiate between viable and dead zoospores using propidium iodide staining and verified that the viability of Bsal zoospores in vitro ranged from 85 – 97%. Lastly, we were able to differentiate between motile and encysted zoospores using flow cytometry by staining the latter life form with Calcoflour White, and verified these counts using florescent microscopy. Our results demonstrate that flow cytometry is effective at enumerating and differentiating Bsal zoospores.
Collaborating Scientists: Debra Miller and Jeanette Wyneken
Drs. Miller and Wyneken have been studying sea turtles for over 20 years. Together they investigate factors impacting hatchling success, such as contaminant exposure and pathogens. Past studies have focused especially on mercury and selenium concentrations in nesting females and their respective hatchlings. Recently, they have turned their focus toward the impact of elevated beach temperatures on hatchling development and nest success (emergence). Suzanne Lenhart (NIMBioS and Math) recently joined the team to begin modeling some of the data that has been collected over the years. A key feature of Wyneken and Miller’s program is to pair biology graduate students with veterinary students to work as teams in the field and the laboratory. This has proven extremely valuable and a truly eye-opening experience by demonstrating to each student the valuable knowledge brought by the other discipline, and how much more they can accomplish by working together. Many of the relationships forged have carried over to their professional careers, continuing research collaborations and co-authoring papers.
Livestock
Collaborating Scientist: Shigetoshi Eda
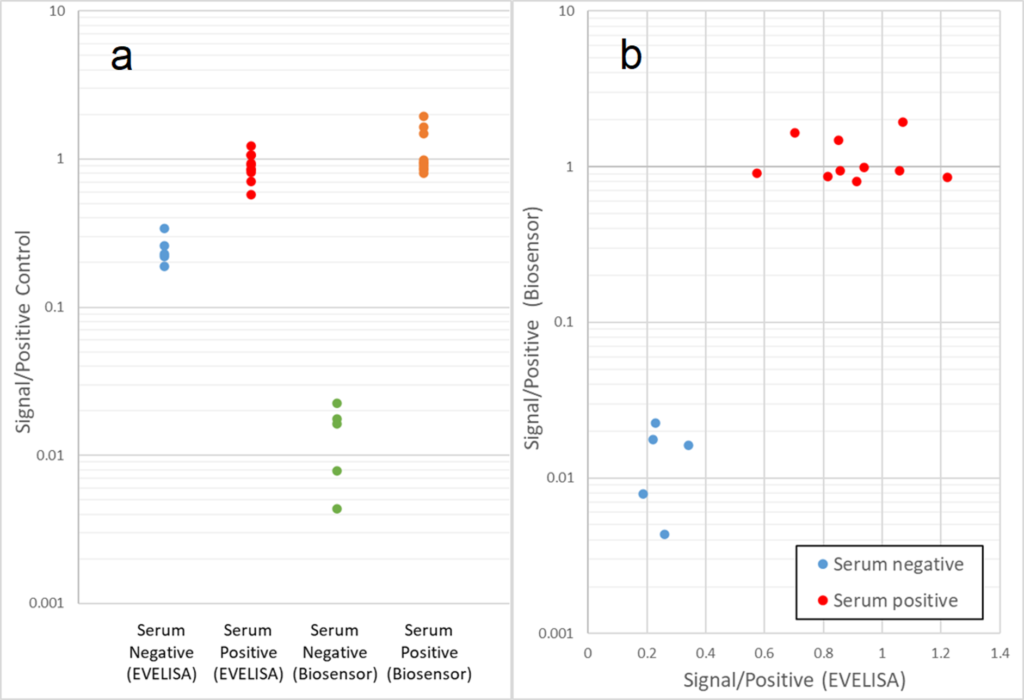
This project entails development of a diagnostic test for Mycobacterium avium subspecies paratuberculosis (Map), which is the causative agent of Johne’s disease (JD). JD causes significant economic losses due to decreased milk production and premature culling of infected animals. Current diagnostic tests are time consuming and/or difficult to operate. This research project aims to develop an on-site method for JD diagnosis employing electrochemically-mediated detection of anti-Map antibodies from sera of JD-affected animals. As a first step toward on-site (lab-on-a-chip) assay development, we confirmed that chromogenic tests utilizing Map antigen-coated magnetic beads could distinguish between JD-positive and negative serum samples. However, since precise optical density measurement requires expensive equipment, we tested if the chromogenic approach could be replaced by electrochemical measurement, by transducing the chemical signal produced using specific redox-active probes. The results of the electrochemical study showed promise that our novel method, which is more sensitive than optical density testing, can be used to differentiate JD-positive and negative serum samples. We are currently testing if the electrochemical detection method can be enhanced through assay procedure modifications.
Mammals
Collaborating Scientists: Melissa Kennedy, Anna McRee, Riley Thompson, Canny Fung, Jessica Dawson, Roger Parry, Chris Foggin, Rebecca Trout-Fryxell, and Agricola Odoi
Project Abstract: Domestic dogs (Canis familiaris) may act as a reservoir for several disease pathogens, including Ehrlichia, which may affect human and wildlife health. Ehrlichia canis is a tick-vectored obligate intracellular bacterium that infiltrates monocytes and macrophages. Blood samples from 225 dogs in northwest Zimbabwe were assessed by serology and 241 samples were assessed by polymerase chain reaction for Ehrlichia. There was a high seroprevalence (73%) of E. canis in domestic dogs in northwest Zimbabwe. After polymerase chain reaction and genetic sequencing, detection of E. canis was reduced (7.5%), indicating that an organism serologically-related to Ehrlichia may be present in the region. This is significant due to the ‘one health’ paradigm, where humans and wildlife may be affected by the high prevalence of this pathogen in domestic dogs.
 UTCVM students processed samples in Zimbabwe. DNA was extracted from whole blood for Ehrlichia PCR.
UTCVM students processed samples in Zimbabwe. DNA was extracted from whole blood for Ehrlichia PCR. UTCVM students processed samples in Zimbabwe. DNA was extracted from whole blood for Ehrlichia PCR.
UTCVM students processed samples in Zimbabwe. DNA was extracted from whole blood for Ehrlichia PCR. UTCVM students prepare blood samples from communal dogs in Zimbabwe.
UTCVM students prepare blood samples from communal dogs in Zimbabwe. UTCVM students collect blood samples from communal dogs in Zimbabwe.
UTCVM students collect blood samples from communal dogs in Zimbabwe. One of Zimbabwe’s communcal dogs.
One of Zimbabwe’s communcal dogs.
Collaborating Scientists: Emma Willcox and Gary McCracken
Graduate Student: Riley Bernard
Project Abstract: Winter emergence of bats from cave hibernacula in the Northeastern US has been attributed to the effects of White-nose Syndrome (WNS). However, our research suggests this may not be the case in the Southeastern US. Our results indicate that in this region, where winters are mild and prey are available, bats regularly leave hibernacula on warm winter nights to forage. Episodic activity and foraging during winter raises body temperature, which should activate the immune system, possibly retarding fungal growth, and resulting in repeated low- level exposures to Pseudogymnoascus destructans (Pd), the causative agent of WNS. The goals of our study are to investigate whether winter cave emergence and foraging behavior contribute to differences in the susceptibility of Pd on bat species known to hibernate in the region. Through non-invasive techniques, our approach will balance research and management by providing potential explanations for
differential Pd susceptibility within and among species and providing management recommendations and mitigation strategies aimed at reducing the effects of WNS and improving winter bat survival.
Background and Significance: Our research suggests that in the Southeastern U.S, where winters are warmer than at more northern latitudes and prey are available, bats are regularly leaving hibernacula to forage. Winter foraging is evidenced by collection of guano samples during capture. Further, we found that more than 50% of bats captured outside hibernacula in Tennessee during the winters of 2012–2013, 2013–2014, and 2014–2015 were negative for Pseudogymnoascus destructans (Pd), the causal agent of WNS, despite coming from infected caves. This indicates Southeastern bats are active outside of caves in winter, regardless of the presence of Pd, and contrasts with evidence from the Northeastern U.S. that suggests activity outside hibernacula during winter is a result of WNS infection. In addition, of the bats we captured that were Pd positive, pathogen load and prevalence varied considerably among species. Episodic activity and foraging during winter raises body temperature, which should activate the immune system possibly retarding fungal growth, and resulting in repeated low-level exposures to the Pd pathogen that could lead to disease immunity. hibernating in southern latitudes with energy not available to more northern bat populations, helping them avoid or fight off Pd infection. Although it is established that bats in the Southeast are active and foraging outside cave hibernacula during winter, the possible impacts on Pd susceptibility and the epizootiology of WNS both within and among species have not been investigated. Differences in winter activity and foraging may account for speciesspecific differences in WNS disease susceptibility, particularly in more southern latitudes, and warrants further investigation. To investigate these issues, we propose examining differences in the winter emergence (activity), and foraging behavior of 3 bat species (Myotis grisescens, M. septentrionalis, and M. sodalis), as a potential explanation for within and among species differences in Pd susceptibility.
Project Goals:
- Assess Pd loads and prevalence in target bat species as an indicator of Pd susceptibility;
- Compare torpor/arousal and winter emergence within and among target bat species including frequency and length of arousal bouts and frequency, duration, and timing of emergence from cave hibernacula
- Contrast foraging behavior among target bat species including location of foraging activity and diet
- Determine the effect torpor, winter emergence, and foraging behavior have on the susceptibility of target bat species to Pd (i.e., Pd loads and prevalence).
Collaborating Scientists: Chunlei Su, Richard Gerhold, and Lisa Muller
Project Abstract: The long-term goal of this study is to elucidate the evolutionary and public health impact of anthropized environment on the transmission of zoonotic pathogens. The burgeoning human population and its increasing demand for food are forcing ever-increasing alterations to natural forests, wetlands, waterways and prairie landscapes. Human activities may profoundly influence zoonotic disease distribution, abundance, and evolution by creating habitats and mediating transmission dynamics among animals that thrive among us, whether as domesticated livestock and companion animals, or as opportunistic pests. Toxoplasma gondii offers us an excellent model to study the impact of anthropized environments on the evolution and transmission of zoonotic pathogens.
This parasite will therefore provide a unique lens through which to understand our indirect, but consequential, influence on parasite ecology, epidemiology, and evolution. The objective of this proposal is to determine the influence of farm cats on T. gondii transmission and
genetic diversity.
Background and Significance: Toxoplasma gondii is transmitted between the definitive host felids (wild and domestic cats), and the intermediate host mammals and birds (Dubey, 2010). It has very little genetic diversity in the north hemisphere but has highly diversity in the south, particularly in South America. In addition, parasite strains in the south hemisphere are more virulent to lab mice, and possibly to humans (Shwab et al., 2014). These attributes of T. gondii are likely influenced by the host species and their environment in different geographical regions. During the past 10,000 years, cats were domesticated as a means to control rodents which became a threat to grain stores in agricultural settlements. However, the unprecedentedly dense populations of mice and cats would then have established an important transmission route for T. gondii.
Project Goals: The goal of this project is to determine if domestic cats cause high contamination of T. gondii oocysts on farms, and facilitate clonal expansion of the parasite.
Collaborating Scientists: Debra Miller, Richard Gerhold, and Chunlei Su
Abstract: Since the discovery of toxoplasmosis in sea otters during a mass die-off in 2006, studies have confirmed the existence of Toxoplasma gondii in many species of marine mammals. However, no cases of toxoplasmosis in manatees were reported until 2012 when 4 Antillean manatees from Puerto Rican waters were confirmed dead due to the disease. This proposed study will use the modified agglutination test (MAT) to determine seroprevalence of T. gondii (i.e., antibodies) in manatees from Puerto Rican waters collected as part of an ongoing capture and release health assessment program led by Dr. Antonio Mignucci-Giannoni (Manatee Conservation Center of Puerto Rico). Additionally, the MAT will be used to test serosanguinous blood pooled from heart, lung, pancreas, and intestinal tissues opportunistically collected from dead manatees from Puerto Rico. The tissues will be examined microscopically for evidence of toxoplasmosis and will also be bioassayed in mice and any organisms retrieved will be genotyped. An additional screening using PCR targeting a T. gondii-specific 529 bp DNA fragment will be performed on individual fresh/frozen tissue samples to further rule-out the presence of organisms within the specific tissue collected. Finally, because felines are the only definitive host of the parasite, serum samples will be collected from feral cats throughout Puerto Rico and tested for T. gondii. Fecal samples will also be collected from the cats and examined microscopically to detect shedding of oocysts. Oocysts that are consistent in size and morphology with T. gondii will be inoculated into mice for propagation and genotyping. Ultimately, the T. gondii genotypes identified from manatees will be compared to those of feral felids. By comparing these genotypes, we hope to establish an epidemiological link to support the hypothesis that feral cat colonies are the source of manatee infection.
Background and Significance: Toxoplasmosis is a parasitic disease common in people, wild and domestic animals worldwide. The parasite has felids as the only definitive host, but can use mammals (especially humans and marine mammals) and birds as intermediate hosts (Nicolle and Manceaux, 1908; Dubey and Frenkel, 1976). Modes of infection include ingestion of sporulated oocysts and tissue cysts, inhalation of aerosolized sporulated oocysts, and transplacental (www.cdc.gov/parasites/toxoplasmosis). The sea otter cases of toxoplasmosis in California in 2002 were linked back to coastal freshwater runoff of feline feces from feral cat populations on the beaches of California (Cole et al., 2000; Miller et al., 2002). Many marine mammals have been the focus of subsequent research; however, surprisingly few studies have been conducted with manatees (Bossart et al., 2012; Dubey et al., 2003), which similar to otters, are coastal species and thus may live in close proximity to feral cat colonies and sewage run-off.
Project Goals: The goal of this project is to investigate the prevalence of T. gondii in Antillean manatees (T. manatus manatus) from Puerto Rico and determine the relatedness of genotypes to those found in Puerto Rican feral cat populations. It will also include research on possible connections between genotypes from the feral cat populations and the genotypes found in the manatees from the same geographical area. The project will indirectly conclude whether an environmental contamination exists. The results of our study will be used in manatee management plans.
Collaborating Scientist: Ashley Reeves, Clayton Hilton, William Swanson, Maria Spriggs
 Figure 1: Semen extraction from a male ocelot being placed into a vial for processing. The ice bags between his legs are to keep him cool on a warm day to avoid heat stress. Anesthetic monitoring is being performed for safety of the animal and personnel.
Figure 1: Semen extraction from a male ocelot being placed into a vial for processing. The ice bags between his legs are to keep him cool on a warm day to avoid heat stress. Anesthetic monitoring is being performed for safety of the animal and personnel. Figure 2: Venipuncture being performed on the jugular of a sedated female ocelot for infectious disease testing. Anesthetic monitoring being performed for the safety of the animal and personnel.
Figure 2: Venipuncture being performed on the jugular of a sedated female ocelot for infectious disease testing. Anesthetic monitoring being performed for the safety of the animal and personnel. Figure 3: Ultrasonography being performed on a sedated female ocelot for determination of pregnancy status.
Figure 3: Ultrasonography being performed on a sedated female ocelot for determination of pregnancy status.
Recent studies concerning genetic variation in wild felid populations have shown that despite captive management, these small populations tend to lose genetic variation over time. The idea of assisted reproductive techniques (ART) have been used in many other species and has the ability to address behavioral or physical incompatibilities between breeding pairs that are genetically suitable, connect regional populations by transporting frozen semen and embryos, preserve genetic diversity with liquid nitrogen tanks, and link wild and zoo populations without bringing more cats into captivity.
Traditionally, semen was collected via electroejaculation and straw sealing for storage in liquid nitrogen vapor. A new method, urethral catheterization, uses alpha-2 agonists such as medetomidine to cause direct sperm release into the urethra. A 3.5 or 5 French X 22 cm urinary catheter is passed 15 cm into the urethra aseptically and held in position for 30 seconds. The catheter is removed, and the semen frozen by two methods: 1) ultra-rapid freezing semen droplets over liquid nitrogen and 2) straw freezing over liquid nitrogen vapor. Post-thaw, the sperm will be assessed for each freezing method for progressive motility over time, intact acrosomes, and in-vitro fertilization of domestic cat oocytes.
Historically, sperm recovered using urethral catheterization had similar total sperm numbers when compared to electroejaculation. In domestic cats, healthy kittens were born following oviductal AI with sperm stored by ultra-rapid freezing. Currently, Dr. Bill Swanson has successfully produced pregnancies by laparoscopic AI in 4 ocelots, among other species of felids using this technique. The main goals of this study are to compare the effectiveness of urethral catheterization using ultra rapid freezing with the traditional straw freezing method in wild ocelots in South Texas, a genetically similar population, to compare results to zoo housed ocelots, genetically variable. Using the ‘Cath-URF’ approach, field veterinarians could opportunistically collect felids in other areas of the world for broader application of ART for felid population management. In addition, females are assessed for pregnancy and vaginal cytology collected for later assessment of the vaginal cells as they relate to current timing of their reproductive cycle. Bobcats are also included in the wild study population as the reproductive capabilities of bobcats have, to our knowledge, never been assessed. Another goal of this study is to determine the prevalence of various infectious diseases including various viral and tick-borne pathogens to compare how the prevalence and incidence of disease has changed in the last 35 years in the ocelot and bobcat populations of South Texas.
Collaborating Scientists: Emma Willcox, Gary McCracken
Graduate Student: Riley Bernard
Project Summary: The gray bat (Myotis grisescens) is 1 of 7 species known to be infected by White-nose syndrome (WNS), a disease caused by the fungal agent Pseudogymnoascus destructans (Pd). As a result of its limited winter distribution and clustering behavior, there were concerns the disease would devastate this species. However, to date, mortality of gray bats from WNS has yet to be reported. Additionally, the results of a previous study, partly funded by a Center for Wildlife Health Seed Grant, suggest Pd loads are significantly lower on active gray bats when compared to numerous other species. We investigated P. destructans fungal loads and WNS infection in torpid and active bats at two large gray bat hibernation sites during the winter of 2014-2015 to try and understand their resistance to the disease. We are currently in the process of determining fungal loads from samples taken from both active and torpid bats.
Results: We have captured and collected Pd swab samples from nearly 200 active bats of 10 species outside cave hibernacula (Pearson and Hubbard’s Caves). 67% of these samples were from active gray bats. In addition, we collected Pd swab samples from 60 torpid gray bats inside the same caves. To date, we have analyzed 152 swab samples for the Pd fungus. Of the 96 samples analyzed from active gray bats, only 8 were Pd positive. Interestingly, only 1 swab taken from the 60 torpid gray bats was Pd positive. If the results of all 152 swab samples so far analyzed are examined, less than 10% were Pd positive, with both loads and prevalence of the fungus lower than in years past. Additional samples collected this winter are currently being analyzed.
Impacts: We will determine the impacts of our project when all of our samples have been fully analyzed and we are able to complete our analyses.
Vectors
Collaborating Scientists: Rebecca Trout Fryxell, Doris D’ Souza, and Suzanne Lenhart
La Crosse encephalitis (LACE) is the most common pediatric arthropod-borne disease in the United States and is an emerging infectious disease in the southern Appalachian region. LACE is caused by infection with the La Crosse virus (LACV), which is transmitted by the bite of infected Aedes mosquitoes. Symptoms usually manifest in children, and complications associated with the disease can be fatal. Because there is no preventive vaccine for LACE, the only effective method for reducing morbidity is the reduction of contact between humans and host-seeking mosquitoes, and because of a lack of sufficient vector control infrastructure, raising community awareness remains the primary method of prevention. As a way to do this, we are collaborating with many residents of east Tennessee (including middle and high school educators) to develop a community-driven mosquito surveillance program. Data obtained through these efforts are used to study the distributions of native and invasive Aedes mosquitoes in order to identify areas with increased risk for LACV transmission where novel small-scale vector control measures may be implemented.
Collaborating Scientists: Rebecca Trout Fryxell, Jennifer DeBruyn, and Steven Wilhelm
Project Abstract: Ticks are hosts to endoymbionts and vectors of pathogens to both humans and animals. As vectors of zoonotic pathogens, ticks acquire their pathogen from an animal host (either wild or domestic) and transmit that pathogen to human and domesticated hosts. Recent studies indicate vector microbiomes influence transmission of vector-borne diseases (i.e., Plasmodium, dengue fever, and trypanosomiasis); however, little is known of Ixodid tick microbiomes. We hypothesize that microbial communities are structured by feeding status and habitat. This is significant because it is speculated that organisms within tick microbiomes may lead to the development of novel disease management strategies as it has for other vector systems. The objectives of this proposal are to define the tick microbiome by characterizing the synergistic, antagonistic, and species-specific bacteria associated within the Lone star tick (Amblyomma americanum) from three different ecoregions (Mississippi Valley Loess Plains, Interior Plateau, and Ridge and Valley) and during three different feeding stages (questing in field, questing on host, and attached). Identifying the microbiota within Lone star ticks known to transmit pathogens and serving as zoonotic vectors can provide an additional option for vector-borne zoonotic disease management to improve human and animal health.
Background and Significance: The vector microbiome can have considerable influence on vector competence (Aksoy 1995, Weis and Aksoy 2011), or the ability of a vector to transmit a pathogen. Recently, a combination of microbiome culturing and sequencing studies has identified bacteria that have significant interactions with their vector-borne pathogen. For example:
- Immune system development and parasite resistance of the tsetse fly are dependent on larvae harboring its endogenous microbiome during intrauterine development (Weis and Aksoy 2011, Weiss et al. 2012). Plasmodium development can be inhibited by bacteria in mosquito midguts, such that increased copies of gram-negative bacteria in Anopheles midguts was associated with lower Plasmodium infection rate and sporogonic-stage development (Pumpuni et al. 1993, Dong et al. 2009, Cirimotich et al. 2011). Eliminating the microbiota within An. gambiae increased the ability of P. falciparum to colonize and replicate within the vector (Dong et al. 2009).
- Chikungunya virus influences the diversity and composition of symbiotic bacteria in colonyraised Ae. albopictus; such that within the vector Alphaproteobacteria, Bacteroidetes and Planctomycetes abundances decreased with increased viral infection (Zouache et al. 2012).
- In colony, when Ae. aegypti harboring field-derived bacterial isolates in the midgut there exhibited a marked decrease in the titre susceptibility of dengue virus (Luis Ramierez et al. 2012).
The role of symbiotic bacteria within vectors is promising as microbes may effectively modulate vector competence; yet very little of this research has been directed on identifying the tick- microbiome, its relation to tick-borne diseases (TBDs), or its association with the environment. Each tick species and life stage is a unique microcosm of microorganisms, some of which are pathogenic to humans (e.g. R. rickettsii and R. parkeri) and some of which are competing with other microbes such as R. amblyommii (a nonpathogenic endosymbiont within Lone star ticks). Our idea stems from the knowledge that while colonizing and replicating within the tick midgut, pathogenic bacteria are competing with a diverse community of microflora (bacteria, fungi, protozoans and viruses) for resources and space (Crotti et al. 2012). Here, we propose to conduct foundational research for tick-habitat/host-microbiome studies.
Project Goals: The goal of this project is to characterize the microbiome of Lone star ticks by describing the microbiomes from different ecoregions and from different feeding statuses to assist with identification of novel bacteria (and eventually mechanisms) for tick management. The results of our study are to provide new perspectives on tick microbiomes as they relate to feeding, habitats, and host choice. Knowledge of the microbial communities within Lone star ticks from different habitats and at different feeding statuses (combined telling a small portion of the life history) will be vital for the predication of both evolutionary responses to current control strategies, and for the development of novel management tactics. We expect to identify microbial signatures associated with specific habitats and/or feeding status.
Collaborating Scientist: Graham Hickling
The Lyme Gradient Project is a collaborative effort between The University of Tennessee, Michigan State University, The University of Montreal, Hofstra University, Georgia Southern University, The University of Rhode Island, and the U.S. Geological Survey. The project aims to understand the ecological drivers for geographic variation in Lyme disease risk in eastern North America. Data on tick-host relationships, tick seasonal biology, and tick genetics are being collected from eight field sites in widely-separated regions of the eastern US. Application of modeling and molecular tools are helping to unravel the ecological and evolutionary processes responsible for the variation in LD risk in different regions of the US, as well as help predict how climate change could alter this risk. Visit the Lyme Gradient website.
Collaborating Scientists: Richard Gerhold, David Buehler, Craig Harper, Roger Applegate, and Roger Shields
Graduate Students: Laura Horton and Sawsan Ammar
There is a perceived decline of wild turkeys (Meleagris gallopavo silvestris) in multiple adjacent counties in south central Tennessee. To determine what factor(s) may be associated with the decline, we have performed a series of experiments and investigations including:
- gross and histological examination of hunter harvested birds in areas of turkey decline as well as areas in which the population seems stable,
- a bioassay study consisting of healthy wild turkeys housed on discarded commercial poultry litter from a poultry farm in the region of the turkey decline to determine if poultry litter can be a source of disease transmission,
- serological testing of turkey serum for exposure to various pathogens, and
- collection and identification of intestinal and ecto parasites.
To date, our research has demonstrated that various pathogens have been detected in wild turkeys collected from hunters and we have found numerous nematode, Cestode, trematode, and protozoal parasites. For the bioassay study, two turkeys developed Blackhead disease due to Histomonas meleagridis, but it was unclear whether H. meleagridis was contracted directly from litter or from ingestion of arthropod pests within the litter. To determine the route of transmission of H. meleagridis, from poultry litter to turkeys, we molecularly tested 387 adult and seven larval darkling beetles (Alphitobius diaperinus) recovered from the poultry litter. Twenty (5.1%) adult or larval beetles were PCR and sequence positive for H. meleagridis. These results indicate that the beetles originating from the poultry litter used in the bioassay study harbored H. meleagridis, either mechanically or internally. We are continuing this health monitoring project in conjunction with a field study directed by Dr. Dave Buehler to examine the biotic and abiotic factors associated with turkey survival and recruitment.

Collaborating Scientists: Richard Gerhold, Shelley Newman, Ed Ramsay, Bob Donnel, and Brad Miller
Graduate Students: Bree Dell and Kate Purple
Echinococcus granulosus (i.e E. canadensis) is a zoonotic cestode parasites in which canids are the definitive hosts shedding the parasite eggs and ungulates are the intermediate hosts consuming the eggs and forming cysts. Few reports of Echinococcus spp. have been described in the US; however, the geographical distribution of Echinococcus spp. in wild hosts is increasing consequent to human activities. In the early 2000’s, 253 elk (Cervus canadensis) originating from Alberta, Canada, were released into the Great Smoky Mountains National Park and North Cumberland Wildlife Management Area in an effort to re-establish their historical range. We investigated the prevalence of Echinococcus spp. in re-established elk populations in the North Cumberland Wildlife Management Area and the Great Smoky Mountains National Park via a retrospective analysis of banked elk tissues and helminth examinations on intestinal contents from coyotes (Canis latrans) from the North Cumberland Wildlife Management Area. Four elk were PCR and sequence positive for E. canadensis. Each sequence had 98% or greater coverage and identity to multiple E. canadensis genotypes in Genbank. Adult Echinococcus spp. were not detected in any of the coyotes examined in this study. Continued surveillance of this disease in susceptible species in these areas is warranted, and these data further underscore the risk of zoonotic pathogen introduction secondary to wildlife translocation. Our laboratory is continuing surveillance for Echinococcus as well as other canid parasites and pathogens in coyotes from eastern Tennessee. We are also working with Tennessee Wildlife Resources Agency to examine carcasses of hunter killed or otherwise killed elk from Tennessee for Echinococcus lesions.
Collaborating Scientists: Richard Gerhold, Chunlei Su, Cheryl Greenacre, and Graham Hickling
Graduate Students: Sawan Ammar, Tania Dawant, Elisa Baker, and Nicole Szafranski
Toxoplasma gondii is a zoonotic protozoal parasite that is the causative agent of Toxoplasmosis. Domestic and wild felids shed the parasite oocysts in their feces and the oocysts are environmentally resistant. All birds and mammals are susceptible to infection with this parasite and infections are lifelong. Susceptible animals are infected by ingesting the microscopic oocysts in food or water or by ingesting contaminated meat that is undercooked or raw. Our laboratory has been studying the ecology and transmission of T. gondii in wildlife using a combination of serological testing in conjunction with high resolution genotyping of the parasites from seropositive animals. To date we have detected T. gondii infections in numerous avian and mammalian species and have shown that there are multiple genotypes circulating in wildlife in the southeastern United States. In addition we have discovered new genotypes in deer, coyotes, waterfowl and wild turkeys. Other studies have shown that urbanization and landscape disturbance are factors for increased T. gondii prevalence, with particular increase in areas with higher domestic cat populations. Our most recent research has demonstrated an unexpectantly high seroprevalence of T. gondii in waterfowl. More research is needed to understand the epidemiology of this infection. Current or upcoming research projects in my laboratory include investigating the tropism, pathology, and survival rate of select waterfowl species experimentally infected with T. gondii, the prevalence of the parasite in wild turkeys, elk, raptors, and white-tailed deer, and the ecosystem contamination of T. gondii by examining the presence of the parasite in soil, water, and invertebrates from select areas we have positive wild mammals or birds. Further publications on our work can be found at the following links.
- Toxoplasma gondii seroprevalence and genotype diversity in select wildlife species from the southeastern United States
- Human impact on the diversity and virulence of the ubiquitous zoonotic parasite Toxoplasma gondii
- A partition of Toxoplasma gondii genotypes across spatial gradients and among host species, and decreased parasite diversity towards areas of human settlement in North America
- Zoonotic Diseases Associated with Free-Roaming Cats
Collaborating Scientists: Richard Gerhold, Stephen Kania, Kim Newkirk, Michelle Dennis, Jullie Ellis, and Krysten Schuler
Graduate Students: Jessie Richards and Abby Wilson
Parelaphostrongylus tenuis (meningeal worm or brain worm) is a parasitic nematode found in various cervid species including deer, elk (Cervus canadensis), and moose (Alces alces). In cervids other than the parasite’s definitive host, white-tailed deer (Odocoileus virginianus), the parasite can migrate extensively in the central nervous system leading to ataxia or other neurological deficiencies and death. Techniques currently available for a definitive diagnosis of the parasite involve a necropsy to detect the worm in the brain and spinal cord via histological exam and/or PCR. Unfortunately this testing is expensive, time consuming and has low sensitivity due to inability to often detect the parasite. Our aim is to develop a rapid, inexpensive, sensitive and specific serological test to detect P. tenuis antibodies in banked or live animal serum. Initially we utilized a previously published peptide.Western blots were utilized to identify anti-P. tenuis antibody present in blood, serum and spinal fluid using sera from known positive animals. However, due to inconsistent results using this protein and cross reactivity with other similar organisms, we moved our attention to a full genomic analysis of P. tenuis which led to identification of a novel P. tenuis-specific peptide. ELISA results utilizing this novel peptide disclosed good reactivity to serum of moose with confirmed P. tenuis infection and minimal reactivity to elk serum from western Wyoming where P. tenuis is not known to occur. We will continue to test known P. tenuis positive sera as well as sera from moose and elk from western US to confirm the sensitivity and specificity of this assay.

Collaborating Scientist: Richard Gerhold and Jane Carlton
Graduate Students: Sawsan Ammar and Kate Purple
The protozoa pathogens Trichomonas gallinae and Histomonas meleagridis are the causative agents of avian trichomonosis (i.e. frounce) and blackhead, respectively. Both of these diseases are emerging issues in both wild and domestic birds. Trichomonosis is considered the most important disease for Mourning Doves and can have variable population impacts on other Columbidae species as well as raptors that feed on sick birds. Recently the disease has been linked to decline of backyard birds in the UK and outbreaks have been reported in the US, Canada, and Scandinavia. These outbreaks have centered on backyard bird baths as a source of infection. Blackhead is mainly a disease of galliforms and wild turkeys are very sensitive to the parasite. Infections in wild turkeys are rapid and can lead to 100% mortality in infected birds. Diagnosing the parasites currently requires sending carcasses to a laboratory for necropsy, PCR, and/or culture. To date, there is no commercial available media for H. meleagridis or T. gallinae and other trichomonads. We have been funded by BioMed Diagnostics to design media that is capable of supporting growth of these avian flagellates which would facilitate the diagnosis of the parasite as well as collection of isolates for research purposes. In addition we are performing molecular testing of these protozoa to understand the molecular epidemiology and transmission. Furthermore, we are also continuing to examine the role that bird baths may play in the spread of T. gallinae and ways to mitigate infection. For more information please see links below.
- Trichomonas gallinae Persistence in Four Water Treatments
- Artificially decreased dissolved oxygen increases the persistence of Trichomonas gallinae in water
- Detection of Trichomonas gallinae in Wild Birds Admitted to a Rehabilitation Center, Florida, USA
- Prevalence and Risk Factors of Trichomonas gallinae and Trichomonosis in Golden Eagle (Aquila chrysaetos) Nestlings in Western North America

Collaborating Scientist: Richard Gerhold and Chunlei Su
Graduate Students: Caroline Grunenwald and Jessie Richards
Moose (Alces alces) are valued species in in many regions. However, the moose populations in multiple regions of North America have experienced declines in their range and population estimates. Nowhere is this decline more pronounced than in Minnesota where the moose population has experienced a sharp, sudden decline including extirpation in the northwest portion of the state and a 66% decline in the last decade in the northeast portions of the state. Although the exact cause of this decline is unclear, parasitic metastrongylid and filarioid nematode infections are known causes of morbidity and mortality in moose across North America. Among the drivers leading to increased parasite prevalence, climate change is considered a major component for both metastrongylid and filarioid nematode. To further assess the impact of parasites in this moose population, we molecularly examined tissue from 34 Minnesota moose that died of known and unknown causes for the presence of nematode DNA. DNA sequencing revealed that PCR products obtained from 15 (44.1%) of the moose were 99% identical to Parelaphostrongylus tenuis, a metastrongylid known to cause neurological disease and death. Additionally, brain tissue from 20 (58.8%) individuals yielded sequences that most closely aligned with Elaeophora schneideri, a parasite associated with neurological impairment but previously unreported in Minnesota. Setaria yehi, a common filarioid parasite of deer, was also detected in the brain tissue of 5 (14.7%) moose. Molecular screening of 618 captured tabanid flies from four trapping sites revealed E. schneideri was present (6%) in the Minnesota environment and transmission could occur locally. These data confirm that P. tenuis is widespread in the Minnesota moose population and raises the question of the significance of E. schneideri as a contributing factor to morbidity and mortality in moose. Our current research is aimed at characterizing the genetic diversity of these parasites to determine transmission dynamics. In addition, we are constructing ELISA tests to detect antibodies to these parasites in the serum of infected animals that will allow us to determine the importance of these parasites in moose health. More information can be found in the publications at the following links:
Ticks and their pathogens are expanding their ranges in North America which includes Tennessee and the southeastern United States. This is a significant concern because ticks transmit an array of pathogens and cause toxic conditions, affecting a variety of host species. Contact between companion animals, livestock, and wildlife is a frequent occurrence and provides opportunities for spillover effects to humans and other animals. We investigate the life histories of ticks and their pathogens to develop prevention, detection, and response strategies for infected-ticks capable of transmitting pathogens or whose blood-feeding behaviors cause expensive economic effects. This is documented with the blacklegged tick (Ixodes scapularis), whose immature life stages feed on hosts such as white-footed mouse or lizards and adult stage feed on white-tailed deer and other larger mammals. Understanding these relationships help to explain why managing mice will curb Lyme disease cases (natural reservoir for Borrelia burgdorferi)and managing deer will curb tick populations.
Recently, the Asian longhorned tick (Haemaphysalis longicornis) was discovered in East Tennessee. This is important because this tick species is an economic threat to agricultural industries (e.g., blood-feeding populations can cause anemia and individuals can transmit a variety of pathogens). Since many tick species are expanding into the region, we are developing a comprehensive tick surveillance program, to detect ticks as soon as possible and use those collections to develop educational material for stakeholders and researchers. In addition, this research aims to identify predictors for detection and assess management options. Understanding the relationships between host life histories, particularly those transfigured due to human behavior, are important considerations for health and the transmission of tick-borne pathogens.
 Removing adult black legged ticks from a white-tailed deer at a wildlife check station.
Removing adult black legged ticks from a white-tailed deer at a wildlife check station. Removing nymph black legged ticks from a white-footed deer mouse after mammal trapping.
Removing nymph black legged ticks from a white-footed deer mouse after mammal trapping.